Getting started: Difference between revisions
| (84 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{latexPreamble}} | {{latexPreamble}} | ||
== Installation == | |||
If you just want a quick try, or if you are not running a linux environment, then you could try [http://www.virtualbox.org virtualbox] and the [[Xubuntu-Openpipeflow_virtual_disk_image]]. | |||
The following few commands are for installation in a linux environment. (On a Mac you could try 'brew install ...' [https://brew.sh/].) | |||
If you've not already, [[download]] the code and unpack the tarball: | |||
:~> tar -xvvzf Openpipeflow-x.xx.tgz | |||
If you have administrator privilages, with linux try | |||
:~> sudo apt-get install gfortran | |||
:~> sudo apt-get install liblapack-dev | |||
:~> sudo apt-get install libfftw3-dev | |||
:~> sudo apt-get install libnetcdf-dev | |||
:~> sudo apt-get install libnetcdff-dev | |||
# optionally, for parallel MPI use: | |||
:~> sudo apt-get install libopenmpi-dev | |||
Then | |||
:~> cd openpipeflow-x.xx/ | |||
:~> make | |||
If this does not end with an error message, then you can skip the section on libraries! Next | |||
:~> make clean | |||
'''NOTE: If you wish to run in parallel, then you will need an mpi-fortran compiler. BUT, the code is sufficiently parallelized that it does NOT require parallel versions of LAPACK, netCDF or FFTW3. Each thread calls the serial implementation of each library.''' | |||
'''Next''' try the [[Tutorial]], or take a look below if there were problems with libraries. | |||
== Overview of files == | == Overview of files == | ||
* '''<tt>Makefile</tt>''' | [[File:Modules4.png]] | ||
* '''<tt>parallel.h</tt>''' Ensure <tt>_Nr</tt> and <tt>_Ns</tt> are both set to <tt>1</tt> if you do not have MPI. This file contains macros for parallelisation that are only invoked if the number of processes <tt>_Np=_Nr*_Ns</tt> is greater than 1. Unless you need many cores, | |||
* '''<tt>Makefile</tt>''' is likely to require modification for your compiler and libraries (see [[#Libraries]]). It has been set up for <tt>gfortran</tt>, but compile flags for several compilers (<tt>g95, gfortran, ifort, pathf90, pgf90</tt>) can be found at the top of the file. | |||
* '''<tt>parallel.h</tt>''' Ensure <tt>_Nr</tt> and <tt>_Ns</tt> are both set to <tt>1</tt> if you do not have MPI. This file contains macros for parallelisation that are only invoked if the number of processes <tt>_Np=_Nr*_Ns</tt> is greater than 1. Unless you need many cores, vary <tt>_Nr</tt> only, keep <tt>_Ns=1</tt> . | |||
* '''<tt>program/parameters.f90</tt>'''. Reynolds number, resolution, timestep, etc. See [[#Parameters]] | * '''<tt>program/parameters.f90</tt>'''. Reynolds number, resolution, timestep, etc. See [[#Parameters]] | ||
* '''<tt>utils/</tt>''' contains a number of utilities. Almost anything can be done in a util, both post-processing and analysing data at runtime. There should be no need to alter the core code. See [[#Making_utils]] | * '''<tt>utils/</tt>''' contains a number of utilities. Almost anything can be done in a util, both post-processing and analysing data at runtime. There should be no need to alter the core code. See [[#Making_utils]] | ||
* '''<tt> | * '''<tt>matlab/</tt>''', a few scripts. See <tt>matlab/Readme.txt</tt> . | ||
== Makefile == | |||
A quick description of the Makefile below. There are suggested flags for several compilers at the top of this file, but the last group (compiler and paths) is the most important section if compiling for the first time. Note the include/link paths specified with <tt>-I<path></tt> and <tt>-L<path></tt> | |||
INSTDIR = ./install/ [no need to change; where to put main.out and main.info files] | |||
PROGDIR = ./program/ [no need to change; where code is located] | |||
UTILDIR = ./utils/ [no need to change; where utility programs are located] | |||
UTIL = prim2matlab [See [[#Making_utils]]; name of utility to build] | |||
TRANSFORM = fftw3 [no need to change; name of FFT library] | |||
MODSOBJ = io.o meshs.o mpi.o parameters.o \ | |||
timestep.o transform.o variables.o velocity.o | |||
#COMPILER = g95 #-C [sample flags for several compilers] | |||
#COMPFLAGS = -cpp -c -O3 | |||
#COMPILER = ifort -i-dynamic #-C #-static | |||
#COMPFLAGS = -cpp -c -O3 -heap-arrays 1024 -mcmodel=medium | |||
#COMPILER = pgf90 #-C | |||
#COMPFLAGS = -Mpreprocess -c -fast #-mcmodel=medium | |||
#COMPILER = pathf90 #-pg -C | |||
#COMPFLAGS = -cpp -c -O3 -OPT:Ofast -march=opteron -fno-second-underscore | |||
COMPILER = gfortran [***CHANGE TO mpif90 FOR PARALLEL USE***] | |||
COMPFLAGS = -ffree-line-length-none -x f95-cpp-input -c -O3 \ | |||
-I/usr/include \ [***MIGHT NEED TO UPDATE INCLUDE PATH***] | |||
#-C #-pg [ See info in following section ] | |||
LIBS = \ | |||
-L/home/ash/lib \ [***MIGHT NEED TO UPDATE LIBRARY PATH***] | |||
cheby.o -lfftw3 -llapack -lnetcdff -lnetcdf \ | |||
# -lblas -lcurl | |||
== Libraries == | |||
'''The code uses freely availably libraries LAPACK, netCDF and FFTW3. If compiling for the first time, consider trying the <tt>gfortran</tt> + ''precompiled binaries'' combination.''' | |||
'''NOTE: The code is sufficiently parallelized that it does not require parallel versions of LAPACK, netCDF or FFTW3. Each thread can calls the serial implementation.''' | |||
In the openpipeflow directory, try <tt>make</tt>, looking for errors indicating missing libraries | |||
> make | |||
... | |||
Fatal Error: Can't open module file 'netcdf.mod' | |||
/usr/bin/ld: cannot find -llapack | |||
/usr/bin/ld: cannot find -lfftw3 | |||
If you get something like | |||
Error: Can't open included file 'mpif.h' | |||
it is possible that <tt>_Np (=_Nr*_Ns)</tt> is not <tt>1</tt> in <tt>parallel.h</tt>, but the <tt>COMPILER</tt> has not been changed to <tt>mpif90</tt> in <tt>Makefile</tt>. | |||
=== Precompiled binaries === | |||
If using a linux environment, install the LAPACK, FFTW3 and netCDF packages with your package manager or 'software center'. If you have sudo privileges, it may be sufficient to try | |||
:~> sudo apt-get install ... | |||
commands listed at the top of this page. Then | |||
:~> cd openpipeflow-x.xx/ | |||
:~> make | |||
If libraries are missing for netCDF, find the location of <tt>netcdf.mod</tt> | |||
:~> cd / | |||
:/> ls */netcdf.mod | |||
:/> ls */*/netcdf.mod | |||
/usr/include/netcdf.mod | |||
Ensure that the file is in the include path in <tt>Makefile</tt>, here <tt>-I/usr/include</tt> | |||
Often the package manager supplies several versions of a package, distinguished by extra extensions. If a library has been installed but is not found, then search for the library, e.g. | |||
:/> ls */*/liblapack* */*/*/libfftw3* */*/*/libnetcdf* | |||
etc/alternatives/liblapack.so.3 | |||
etc/alternatives/liblapack.so.3gf | |||
usr/lib/i386-linux-gnu/libfftw3_omp.so.3 | |||
usr/lib/i386-linux-gnu/libfftw3_omp.so.3.3.2 | |||
usr/lib/i386-linux-gnu/libfftw3.so.3 | |||
usr/lib/i386-linux-gnu/libfftw3.so.3.3.2 | |||
usr/lib/i386-linux-gnu/libnetcdff.so.6 | |||
usr/lib/i386-linux-gnu/libnetcdf.so.11 | |||
Create a symbolic link to one of the versions for each package | |||
:~> mkdir /home/ash/lib | |||
:~> ln -s /etc/alternatives/liblapack.so.3 /home/ash/lib/liblapack.so | |||
:~> ln -s /usr/lib/i386-linux-gnu/libfftw3.so.3 /home/ash/lib/libfftw3.so | |||
:~> ln -s /usr/lib/i386-linux-gnu/libnetcdff.so.6 /home/ash/lib/libnetcdff.so | |||
:~> ln -s /usr/lib/i386-linux-gnu/libnetcdf.so.11 /home/ash/lib/libnetcdf.so | |||
Ensure that they are in the link path in <tt>Makefile</tt>, <tt>-L/home/ash/lib</tt> | |||
'''Note:''' Some extra libraries may or may not be needed, depending on compiler/distribution. Try simply omitting the link flags | |||
<tt>-lblas</tt> and/or <tt>-lcurl</tt> at first, for example. | |||
=== Compiling libraries === | |||
If it is not possible to link with precompiled libraries, or if special flags are necessary, e.g. for a very large memory model, | |||
then it may be necessary to build LAPACK and NetCDF with the compiler and compile flags that will be used for the main simulation code. | |||
The default procedure for building a package (FFTW3 | The default procedure for building a package (applicable to FFTW3 and netCDF) is | ||
<pre>tar -xvvzf package.tar.gz | <pre>tar -xvvzf package.tar.gz | ||
cd package/ | cd package/ | ||
[set environment variables if necessary] | [set environment variables if necessary] | ||
./configure --prefix=<path> | ./configure --prefix=<path to put libraries> | ||
make | make | ||
make install</pre> | make install</pre> | ||
| Line 29: | Line 143: | ||
<tt>cp blas.a <path>/lib/libblas.a</tt> | <tt>cp blas.a <path>/lib/libblas.a</tt> | ||
– '''netCDF'''. If using gfortran you might be able to use the supplied binary version for your linux distribution. Several versions can be found at <tt>http://www.unidata.ucar.edu/downloads/netcdf/current/index.jsp</tt>. Version '''4.1.3''' is relatively straight forward to install, the following typical environment variables required to build netCDF should be sufficient: | – '''netCDF'''. If using gfortran you might be able to use the supplied binary version for your linux distribution. Several versions can be found at <tt>http://www.unidata.ucar.edu/downloads/netcdf/current/index.jsp</tt>. | ||
Version '''4.1.3''' is relatively straight forward to install, the following typical environment variables required to build netCDF should be sufficient: | |||
<br /> | <br /> | ||
<tt>CXX=""</tt><br /> | <tt>CXX=""</tt><br /> | ||
| Line 37: | Line 153: | ||
After building, ensure that files <tt>netcdf.mod</tt> and <tt>typesizes.mod</tt> appear in your include path (see Makefile COMPFLAGS -I$<$path$>$/include/). | After building, ensure that files <tt>netcdf.mod</tt> and <tt>typesizes.mod</tt> appear in your include path (see Makefile COMPFLAGS -I$<$path$>$/include/). | ||
netCDF installation is slightly trickier | For more recent versions, netCDF installation is slightly trickier [currently netcdf-4.3.0.tar.gz 2015-07-20]. First<br /> | ||
<tt>./configure --disable-netcdf-4 --prefix | <tt>./configure --disable-netcdf-4 --prefix=<path></tt><br /> | ||
which disables HDF5 support (not currently required, see comment on [[Parallel_i/o]]). Also, Fortran is no longer bundled, so get netcdf-fortran-4.2.2.tar.gz or a more recent version from here<br /> | which disables HDF5 support (not currently required, see comment on [[Parallel_i/o]]). Also, Fortran is no longer bundled, so get netcdf-fortran-4.2.2.tar.gz or a more recent version from here<br /> | ||
<tt>http://www.unidata.ucar.edu/downloads/netcdf/index.jsp</tt><br /> | <tt>http://www.unidata.ucar.edu/downloads/netcdf/index.jsp</tt><br /> | ||
| Line 51: | Line 167: | ||
<tt> _Nr</tt> split in radius<br /> | <tt> _Nr</tt> split in radius<br /> | ||
<tt> _Ns</tt> split axially<br /> | <tt> _Ns</tt> split axially<br /> | ||
Number of | Number of cores is <tt>_Np = _Nr * _Ns</tt>. | ||
'''Set both <tt>_Nr</tt> and <tt> | '''Set both <tt>_Nr</tt> and <tt>_Ns</tt> to <tt>1</tt> for serial use. MPI not required in that case.''' | ||
Unless a large number of cores is required, it is recommended to vary only <tt>_Nr</tt> and to keep <tt>_Ns=1</tt>, i.e. split radially only. Prior to openpipeflow-1.10, only <tt>_Np</tt> was available, equivalent to the current <tt>_Nr</tt>. | Unless a large number of cores is required, it is recommended to vary only <tt>_Nr</tt> and to keep <tt>_Ns=1</tt>, i.e. split radially only. Prior to openpipeflow-1.10, only <tt>_Np</tt> was available, equivalent to the current <tt>_Nr</tt>. | ||
| Line 97: | Line 213: | ||
<tr class="odd"> | <tr class="odd"> | ||
<td align="left"><tt>i_save_rate1</tt></td> | <td align="left"><tt>i_save_rate1</tt></td> | ||
<td align="left">Save frequency for snapshot data files</td> | <td align="left">Save frequency for snapshot data files (timesteps between saves)</td> | ||
</tr> | </tr> | ||
<tr class="even"> | <tr class="even"> | ||
| Line 175: | Line 291: | ||
<td align="left"><tt>vel_energy.dat</tt></td> | <td align="left"><tt>vel_energy.dat</tt></td> | ||
<td align="left">$t$, $E$, $E_{k=0}$, $E_{m=0}$</td> | <td align="left">$t$, $E$, $E_{k=0}$, $E_{m=0}$</td> | ||
<td align="left">energies. $E-E_{k=0} | <td align="left">energies. Streamwise-dependent component of energy = $E-E_{k=0}$.</td> | ||
</tr> | </tr> | ||
<tr class="odd"> | <tr class="odd"> | ||
| Line 181: | Line 297: | ||
<td align="left">$t$, $U_b$ or $\beta$, $\langle u_z(r=0)\rangle_z$, $u_\tau$</td> | <td align="left">$t$, $U_b$ or $\beta$, $\langle u_z(r=0)\rangle_z$, $u_\tau$</td> | ||
<td align="left">bulk speed or pressure measure $1+\beta=Re/Re_m$, mean centreline speed, friction vel.</td> | <td align="left">bulk speed or pressure measure $1+\beta=Re/Re_m$, mean centreline speed, friction vel.</td> | ||
</tr> | |||
<tr class="even"> | |||
<td align="left"><tt>vel_totEID.dat</tt></td> | |||
<td align="left">$t$, $E/E_{lam}$, $I/I_{lam}$, $D/D_{lam}$</td> | |||
<td align="left">total energy, input, dissipation; each normalised by laminar value.</td> | |||
</tr> | </tr> | ||
</table> | </table> | ||
| Line 186: | Line 307: | ||
== Typical usage == | == Typical usage == | ||
'''See | '''See the [[Tutorial]] for the fundamentals of setting up, launching, monitoring and ending a job.''' | ||
==== Serial and parallel use ==== | |||
* To launch a serial job | |||
> nohup ./main.out > OUT 2> OUT.err & | |||
* The number of cores is set in <tt>parallel.h</tt> (all other parameters are in <tt>program/parameters.f90</tt>). | |||
* For serial use, both <tt>_Nr</tt> and <tt>_Ns</tt> should be <tt>1</tt> in <tt>parallel.h</tt>. | |||
* | * For parallel use, in <tt>Makefile</tt> the <tt>COMPILER</tt> needs to be changed to e.g. <tt>mpif90</tt>. | ||
* MPI is required if, and only if, <tt>_Np=_Nr*_Ns</tt> is greater than <tt>1</tt> in <tt>parallel.h</tt>. | |||
> | * For modest parallelisation, <tt>_Np</tt><=32, it is recommended to change <tt>_Nr</tt> only. Leave <tt>_Ns</tt> set to <tt>1</tt> (i.e. <tt>_Np==_Nr</tt>). | ||
* For high parallelisation, <tt>_Nr</tt> and <tt>_Ns</tt> should be of the same order. | |||
* To launch a parallel job on 8 cores | |||
> nohup mpirun -np 8 ./main.out > OUT 2> OUT.err & | |||
* Note that, rather than keep changing <tt>COMPILER</tt> when switching between parallel and serial use, it may be more convenient to simply launch with <tt>-np 1</tt> in the parallel launch command. | |||
==== Initial conditions ==== | |||
The best initial condition is the state with most similar parameters. Some are supplied in the [[Database]]. | |||
Any output state, e.g. state0012.cdf.dat can be copied to <tt>state.cdf.in</tt> to be used as an initial condition. If resolutions do not match, they are '''automatically interpolated or truncated'''. If none is appropriate, then, it may be necessary to create a utility. See [[Utilities]]. | |||
=== | ==== Typical setup commands ==== | ||
> nano program/parameters.f90 [set up parameters and compile] | |||
> make | > make | ||
> make install | > make install | ||
> [prepare a directory for the job] | |||
> | > mkdir ~/runs | ||
> mv install ~/runs/job0009 | |||
> cd ~/runs/job0009 | |||
> | > [get input state.cdf.in from somewhere... e.g. another job] | ||
> | > diff ../job0008/main.info main.info [compare parameters with a similar job - a good check] | ||
> | > cp ../job0008/state0052.cdf.dat state.cdf.in | ||
> [or get a state.cdf.in from | |||
> http://www.openpipeflow.org/index.php?title=Database] | |||
> | |||
> [Run!...] | |||
> nohup ./main.out > OUT 2> OUT.err & [run serial] | |||
> nohup mpirun -np 8 ./main.out > OUT 2> OUT.err & [run parallel] | |||
> | |||
> head OUT [check for warnings/errors] | |||
> tail OUT [see how far job has got] | |||
> | |||
> rm RUNNING [end the job cleanly] | |||
If a job does not launch correctly, the problem can usually be found in the output | |||
> less OUT ['q' to quit] | |||
> less OUT.err | |||
== | == Data processing == | ||
Almost anything can be done either at runtime or as post-processing by creating a utility. | |||
It very rare that the core code in <tt>program/</tt> should need to be changed. | |||
There are many examples in <tt>utils/</tt>. Further information can be found on the [[Utilities]] page. | There are many examples in <tt>utils/</tt>. Further information can be found on the [[Utilities]] page. | ||
Latest revision as of 09:23, 3 December 2024
$ \renewcommand{\vec}[1]{ {\bf #1} } \newcommand{\bnabla}{ \vec{\nabla} } \newcommand{\Rey}{Re} \def\vechat#1{ \hat{ \vec{#1} } } \def\mat#1{#1} $
Installation
If you just want a quick try, or if you are not running a linux environment, then you could try virtualbox and the Xubuntu-Openpipeflow_virtual_disk_image.
The following few commands are for installation in a linux environment. (On a Mac you could try 'brew install ...' [1].)
If you've not already, download the code and unpack the tarball:
:~> tar -xvvzf Openpipeflow-x.xx.tgz
If you have administrator privilages, with linux try
:~> sudo apt-get install gfortran :~> sudo apt-get install liblapack-dev :~> sudo apt-get install libfftw3-dev :~> sudo apt-get install libnetcdf-dev :~> sudo apt-get install libnetcdff-dev # optionally, for parallel MPI use: :~> sudo apt-get install libopenmpi-dev
Then
:~> cd openpipeflow-x.xx/ :~> make
If this does not end with an error message, then you can skip the section on libraries! Next
:~> make clean
NOTE: If you wish to run in parallel, then you will need an mpi-fortran compiler. BUT, the code is sufficiently parallelized that it does NOT require parallel versions of LAPACK, netCDF or FFTW3. Each thread calls the serial implementation of each library.
Next try the Tutorial, or take a look below if there were problems with libraries.
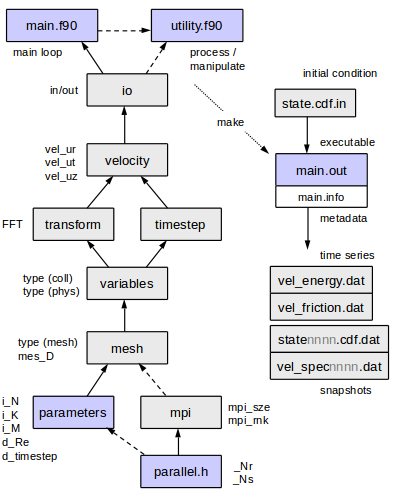
Overview of files
- Makefile is likely to require modification for your compiler and libraries (see #Libraries). It has been set up for gfortran, but compile flags for several compilers (g95, gfortran, ifort, pathf90, pgf90) can be found at the top of the file.
- parallel.h Ensure _Nr and _Ns are both set to 1 if you do not have MPI. This file contains macros for parallelisation that are only invoked if the number of processes _Np=_Nr*_Ns is greater than 1. Unless you need many cores, vary _Nr only, keep _Ns=1 .
- program/parameters.f90. Reynolds number, resolution, timestep, etc. See #Parameters
- utils/ contains a number of utilities. Almost anything can be done in a util, both post-processing and analysing data at runtime. There should be no need to alter the core code. See #Making_utils
- matlab/, a few scripts. See matlab/Readme.txt .
Makefile
A quick description of the Makefile below. There are suggested flags for several compilers at the top of this file, but the last group (compiler and paths) is the most important section if compiling for the first time. Note the include/link paths specified with -I<path> and -L<path>
INSTDIR = ./install/ [no need to change; where to put main.out and main.info files] PROGDIR = ./program/ [no need to change; where code is located] UTILDIR = ./utils/ [no need to change; where utility programs are located] UTIL = prim2matlab [See #Making_utils; name of utility to build]
TRANSFORM = fftw3 [no need to change; name of FFT library]
MODSOBJ = io.o meshs.o mpi.o parameters.o \
timestep.o transform.o variables.o velocity.o
#COMPILER = g95 #-C [sample flags for several compilers] #COMPFLAGS = -cpp -c -O3 #COMPILER = ifort -i-dynamic #-C #-static #COMPFLAGS = -cpp -c -O3 -heap-arrays 1024 -mcmodel=medium #COMPILER = pgf90 #-C #COMPFLAGS = -Mpreprocess -c -fast #-mcmodel=medium #COMPILER = pathf90 #-pg -C #COMPFLAGS = -cpp -c -O3 -OPT:Ofast -march=opteron -fno-second-underscore
COMPILER = gfortran [***CHANGE TO mpif90 FOR PARALLEL USE***]
COMPFLAGS = -ffree-line-length-none -x f95-cpp-input -c -O3 \
-I/usr/include \ [***MIGHT NEED TO UPDATE INCLUDE PATH***]
#-C #-pg [ See info in following section ]
LIBS = \
-L/home/ash/lib \ [***MIGHT NEED TO UPDATE LIBRARY PATH***]
cheby.o -lfftw3 -llapack -lnetcdff -lnetcdf \
# -lblas -lcurl
Libraries
The code uses freely availably libraries LAPACK, netCDF and FFTW3. If compiling for the first time, consider trying the gfortran + precompiled binaries combination.
NOTE: The code is sufficiently parallelized that it does not require parallel versions of LAPACK, netCDF or FFTW3. Each thread can calls the serial implementation.
In the openpipeflow directory, try make, looking for errors indicating missing libraries
> make ... Fatal Error: Can't open module file 'netcdf.mod' /usr/bin/ld: cannot find -llapack /usr/bin/ld: cannot find -lfftw3
If you get something like
Error: Can't open included file 'mpif.h'
it is possible that _Np (=_Nr*_Ns) is not 1 in parallel.h, but the COMPILER has not been changed to mpif90 in Makefile.
Precompiled binaries
If using a linux environment, install the LAPACK, FFTW3 and netCDF packages with your package manager or 'software center'. If you have sudo privileges, it may be sufficient to try
:~> sudo apt-get install ...
commands listed at the top of this page. Then
:~> cd openpipeflow-x.xx/ :~> make
If libraries are missing for netCDF, find the location of netcdf.mod
:~> cd / :/> ls */netcdf.mod :/> ls */*/netcdf.mod /usr/include/netcdf.mod
Ensure that the file is in the include path in Makefile, here -I/usr/include
Often the package manager supplies several versions of a package, distinguished by extra extensions. If a library has been installed but is not found, then search for the library, e.g.
:/> ls */*/liblapack* */*/*/libfftw3* */*/*/libnetcdf* etc/alternatives/liblapack.so.3 etc/alternatives/liblapack.so.3gf usr/lib/i386-linux-gnu/libfftw3_omp.so.3 usr/lib/i386-linux-gnu/libfftw3_omp.so.3.3.2 usr/lib/i386-linux-gnu/libfftw3.so.3 usr/lib/i386-linux-gnu/libfftw3.so.3.3.2 usr/lib/i386-linux-gnu/libnetcdff.so.6 usr/lib/i386-linux-gnu/libnetcdf.so.11
Create a symbolic link to one of the versions for each package
:~> mkdir /home/ash/lib :~> ln -s /etc/alternatives/liblapack.so.3 /home/ash/lib/liblapack.so :~> ln -s /usr/lib/i386-linux-gnu/libfftw3.so.3 /home/ash/lib/libfftw3.so :~> ln -s /usr/lib/i386-linux-gnu/libnetcdff.so.6 /home/ash/lib/libnetcdff.so :~> ln -s /usr/lib/i386-linux-gnu/libnetcdf.so.11 /home/ash/lib/libnetcdf.so
Ensure that they are in the link path in Makefile, -L/home/ash/lib
Note: Some extra libraries may or may not be needed, depending on compiler/distribution. Try simply omitting the link flags -lblas and/or -lcurl at first, for example.
Compiling libraries
If it is not possible to link with precompiled libraries, or if special flags are necessary, e.g. for a very large memory model, then it may be necessary to build LAPACK and NetCDF with the compiler and compile flags that will be used for the main simulation code.
The default procedure for building a package (applicable to FFTW3 and netCDF) is
tar -xvvzf package.tar.gz cd package/ [set environment variables if necessary] ./configure --prefix=<path to put libraries> make make install
– FFTW3. This usually requires no special treatment. Install with your package manager or build with the default settings.
– LAPACK. If using gfortran you might be able to use the binary version supplied for your linux distribution. Otherwise, edit the file make.inc that comes with LAPACK, setting the fortran compiler and flags to those you plan to use. Type ‘make’. Once finished, copy the following binaries into your library path (see Makefile LIBS -L$<$path$>$/lib/)
cp lapack.a <path>/lib/liblapack.a
cp blas.a <path>/lib/libblas.a
– netCDF. If using gfortran you might be able to use the supplied binary version for your linux distribution. Several versions can be found at http://www.unidata.ucar.edu/downloads/netcdf/current/index.jsp.
Version 4.1.3 is relatively straight forward to install, the following typical environment variables required to build netCDF should be sufficient:
CXX=""
FC=/opt/intel/fc/10.1.018/bin/ifort
FFLAGS="-O3 -mcmodel=medium"
export CXX FC FFLAGS
After building, ensure that files netcdf.mod and typesizes.mod appear in your include path (see Makefile COMPFLAGS -I$<$path$>$/include/).
For more recent versions, netCDF installation is slightly trickier [currently netcdf-4.3.0.tar.gz 2015-07-20]. First
./configure --disable-netcdf-4 --prefix=<path>
which disables HDF5 support (not currently required, see comment on Parallel_i/o). Also, Fortran is no longer bundled, so get netcdf-fortran-4.2.2.tar.gz or a more recent version from here
http://www.unidata.ucar.edu/downloads/netcdf/index.jsp
Build with the environment variables above, and in addition
CPPFLAGS=-I<path>/include
export CPPFLAGS
Finally, add the link flag -lnetcdff immediately before -lnetcdf in your Makefile.
Parameters
./parallel.h:
_Nr split in radius
_Ns split axially
Number of cores is _Np = _Nr * _Ns.
Set both _Nr and _Ns to 1 for serial use. MPI not required in that case.
Unless a large number of cores is required, it is recommended to vary only _Nr and to keep _Ns=1, i.e. split radially only. Prior to openpipeflow-1.10, only _Np was available, equivalent to the current _Nr.
_Nr is 'optimal' if it is a divisor of i_N or only slightly larger than a divisor. To keep code simple, _Ns must divide both i_Z and i_M parameters below (easier to leave _Ns=1 if not needed).
./program/parameters.f90:
| i_N | Number of radial points $n\in[1,N]$ |
| i_K | Maximum k (axial), $k\in(-K,\,K)$ |
| i_M | Maximum m (azimuthal), $m\in[0,\,M)$ |
| i_Mp | Azimuthal periodicity, i.e. $m=0,M_p,2M_p,\dots,(M-1)M_p$ |
| (set =1 for no symmetry assumption) | |
| d_Re | Reynolds number $Re$ or $Rm_m$ |
| d_alpha | Axial wavenumber $\alpha=2\pi/L_z$ |
| b_const_flux | Enforce constant flux $U_b=\frac1{2}$. |
| i_save_rate1 | Save frequency for snapshot data files (timesteps between saves) |
| i_save_rate2 | Save frequency for time-series data |
| i_maxtstep | Maximum number of timesteps (no limit if set =-1) |
| d_cpuhours | Maximum number of cpu hours |
| d_time | Start time (taken from state.cdf.in if set =-1d0) |
| d_timestep | Fixed timestep (typically =0.01d0 or dynamically controlled if set =-1d0) |
| d_dterr | Maximum corrector norm, $\|f_{corr}\|$ (typically =1d-5 or set =1d1 to avoid extra corrector iterations) |
| d_courant | Courant number $\mathrm{C}$ (unlikely to need changing) |
| d_implicit | Implicitness $c$ (unlikely to need changing) |
Note the default cases, usually if the parameter is set to -1.
Input files
State files are stored in the NetCDF data format are binary yet can be transferred across different architectures safely. The program main.out runs with the compiled parameters (see main.info) but will load states of other truncations. For example, an output state file state0018.cdf.dat can be copied to an input state.cdf.in, and when loaded it will be interpolated if necessary.
Sample initial conditions are available in the Database.
state.cdf.in:
$t$ – Start time. Overridden by d_time if d_time$\ge$0
$\Delta t$ – Timestep. Ignored, see parameter d_timestep.
$N, M_p, r_n$ – Number of radial points, azimuthal periodicity, radial values of the input state.
– If the input radial points differ from the runtime points, then the fields are interpolated onto the new points automatically.
$u_r,\,u_\theta,\, u_z$ – Field vectors.
– If $K\ne\,$i_K or $M\ne\,$i_M, then Fourier modes are truncated or zeros appended.
Output
Snapshot data
Data saved every i_save_rate1 timesteps:
state????.cdf.dat vel_spec????.dat
All output is sent to the current directory, and ???? indicates numbers 0000, 0001, 0002,…. Each state file can be copied to a state.cdf.in should a restart be necessary. To list times $t$ for each saved state file,
> grep state OUT
The spectrum files are overwritten each save as they are retrievable from the state data. To verify sufficient truncation, a quick profile of the energy spectrum can be plotted with
gnuplot> set log
gnuplot> plot 'vel_spec0002.dat' w lp
Time-series data
Data saved every i_save_rate2 timesteps:
| tim_step.dat | $t$, $\Delta t$, $\Delta t_{\|f\|}$, $\Delta t_{CFL}$ | current and limiting step sizes |
| vel_energy.dat | $t$, $E$, $E_{k=0}$, $E_{m=0}$ | energies. Streamwise-dependent component of energy = $E-E_{k=0}$. |
| vel_friction.dat | $t$, $U_b$ or $\beta$, $\langle u_z(r=0)\rangle_z$, $u_\tau$ | bulk speed or pressure measure $1+\beta=Re/Re_m$, mean centreline speed, friction vel. |
| vel_totEID.dat | $t$, $E/E_{lam}$, $I/I_{lam}$, $D/D_{lam}$ | total energy, input, dissipation; each normalised by laminar value. |
Typical usage
See the Tutorial for the fundamentals of setting up, launching, monitoring and ending a job.
Serial and parallel use
- To launch a serial job
> nohup ./main.out > OUT 2> OUT.err &
- The number of cores is set in parallel.h (all other parameters are in program/parameters.f90).
- For serial use, both _Nr and _Ns should be 1 in parallel.h.
- For parallel use, in Makefile the COMPILER needs to be changed to e.g. mpif90.
- MPI is required if, and only if, _Np=_Nr*_Ns is greater than 1 in parallel.h.
- For modest parallelisation, _Np<=32, it is recommended to change _Nr only. Leave _Ns set to 1 (i.e. _Np==_Nr).
- For high parallelisation, _Nr and _Ns should be of the same order.
- To launch a parallel job on 8 cores
> nohup mpirun -np 8 ./main.out > OUT 2> OUT.err &
- Note that, rather than keep changing COMPILER when switching between parallel and serial use, it may be more convenient to simply launch with -np 1 in the parallel launch command.
Initial conditions
The best initial condition is the state with most similar parameters. Some are supplied in the Database. Any output state, e.g. state0012.cdf.dat can be copied to state.cdf.in to be used as an initial condition. If resolutions do not match, they are automatically interpolated or truncated. If none is appropriate, then, it may be necessary to create a utility. See Utilities.
Typical setup commands
> nano program/parameters.f90 [set up parameters and compile] > make > make install > [prepare a directory for the job] > mkdir ~/runs > mv install ~/runs/job0009 > cd ~/runs/job0009 > [get input state.cdf.in from somewhere... e.g. another job] > diff ../job0008/main.info main.info [compare parameters with a similar job - a good check] > cp ../job0008/state0052.cdf.dat state.cdf.in > [or get a state.cdf.in from > http://www.openpipeflow.org/index.php?title=Database] > > [Run!...] > nohup ./main.out > OUT 2> OUT.err & [run serial] > nohup mpirun -np 8 ./main.out > OUT 2> OUT.err & [run parallel] > > head OUT [check for warnings/errors] > tail OUT [see how far job has got] > > rm RUNNING [end the job cleanly]
If a job does not launch correctly, the problem can usually be found in the output
> less OUT ['q' to quit] > less OUT.err
Data processing
Almost anything can be done either at runtime or as post-processing by creating a utility. It very rare that the core code in program/ should need to be changed.
There are many examples in utils/. Further information can be found on the Utilities page.