Tutorial: Difference between revisions
mNo edit summary |
m (→Start the run) |
||
| (61 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
This tutorial aims to guide a new user through | |||
* setting up parameters for a job, | |||
* monitoring a job's progress, | |||
* simple plotting of time series from a run, | |||
* visualisation of structures in a snapshot, | |||
* manipulating data. | |||
It is advised that you read through the [[Getting_started]] before trying the tutorial, but you may skip the | |||
[[Getting_started#Libraries|Libraries]] section if someone has set up the libraries and <tt>Makefile</tt> for you. | |||
The following assumes that the command '<tt>make</tt>' does not exit with an error, i.e. that the code has been downloaded ([[download]]), and that libraries have been correctly installed ([[Getting_started#Compiling_libraries]]). | |||
=== Overview === | |||
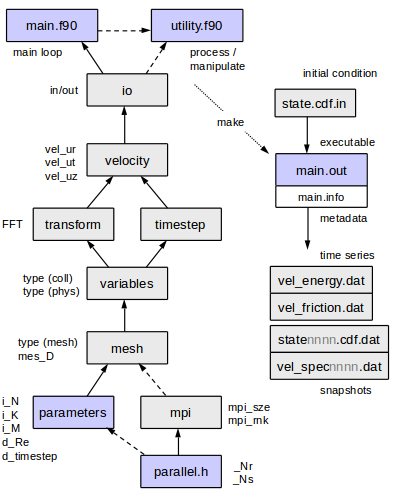
[[File:Modules4.png]] | |||
=== Where to start - initial conditions and the main.info file === | |||
The best input initial condition, <tt>state.cdf.in</tt> is usually the output state from another run, preferably from a run with similar parameter settings. | |||
Output state files are named <tt>state0000.cdf.dat, state0001.cdf.dat, state0002.cdf.dat,</tt> and so on. Any of these could be used as a potential initial condition. If resolution parameters do not match, then they automatically interpolated or truncated to the new resolution (the resolution selected at compile time). | |||
In this example we'll use a travelling wave solution at Re=2400 as an initial condition for a run at Re=2500. Download the following file from the Database: [[File:Re2400a1.25.tgz]] and extract the contents: | |||
> tar -xvvzf Re2400a1.25.tgz | |||
> cd Re2400a1.25/ | |||
> ls -l | |||
-rw-rw-r-- 1 758 Jul 16 2014 main.info | |||
-rw-rw-r-- 1 3177740 Jun 10 2013 state0010.cdf.dat | |||
The directory <tt>Re2400a1.25/</tt> contains an output state file <tt>state0010.cdf.dat</tt> and a <tt>main.info</tt> file. The [[main.info]] file is a plain text record of parameter settings that were used when compiling the executable that produced the state file. | |||
'''A more computationally demanding example:''' You may wish to start from [[File:Re5300.Retau180.5D.tgz]] and compare data with Eggels et al. (1994) JFM. To run at a reasonable pace, try running on 5-10 cores. | |||
=== Set your parameters === | |||
In a terminal window, take a look at the main.info file downloaded a moment ago | |||
> less Re2400a1.25/main.info | |||
('less' is like 'more', but you can scroll and search (with '/'). Press 'q' to exit.) | |||
The [[main.info]] file records the parameters associated with the state file, which will be used as the initial condition. | |||
In another terminal window, edit the [[Getting_started#Parameters|parameters]] for the run, so that they are the same as in the given <tt>main.info</tt> file, except with Re=2500. (If you have not yet changed the <tt>parameters.f90</tt> file, it'll not need editing.) | |||
> nano program/parameters.f90 [OR] | |||
> pico program/parameters.f90 [OR] | |||
> gedit program/parameters.f90 | |||
In the <tt>parameters.f90</tt> file, you should ignore from where <tt>i_KL</tt> is declared onwards. | |||
We will assume serial use (for parallel use see [[Getting_started#Typical_usage]]). | |||
The number of cores is set in <tt>parallel.h</tt>. Ensure that <tt>_Nr</tt> and <tt>_Ns</tt> are both defined to be <tt>1</tt> | |||
(number of cores <tt>_Np=_Nr*_Ns</tt>): | |||
> head parallel.h | |||
... | |||
#define _Nr 1 | |||
#define _Ns 1 | |||
... | |||
If not, edit with your favourite text editor, e.g. | |||
> nano parallel.h | |||
and set <tt>_Nr</tt> and <tt>_Ns</tt> as above. | |||
=== Compile and setup a job directory === | |||
After setting the parameters, we need to create an executable that will run with the settings we've chosen. To '''compile''' the code with the current parameter settings | |||
> make | |||
> make install | |||
If an error is produced, see the top of this page - check that libraries and <tt>Makefile</tt> are set up correctly. | |||
The second command creates the directory <tt>install/</tt> and a new [[main.info]] file. Often its a good idea to check for | |||
any differences in parameters between jobs: | |||
> diff install/main.info Re2400a1.25/main.info | |||
In this case the important difference is Re=2400 increased to Re=2500. | |||
We'll create a new job directory with an initial condition in there ready for the new run | |||
> cp Re2400a1.25/state0010.cdf.dat install/state.cdf.in | |||
> mkdir ~/runs/ | |||
> mv install ~/runs/job0001 | |||
> cd ~/runs/job0001 | |||
> ls -l | |||
-rw-rw-r-- 1 949 Sep 9 08:50 main.info | |||
-rw-rw-r-- 1 3679640 Sep 9 08:50 main.out | |||
-rw-rw-r-- 1 3177740 Jun 10 2013 state.cdf.in | |||
=== Start the run === | |||
To start the run | |||
> nohup ./main.out > OUT 2> OUT.err & | |||
'&' puts the job in the background. | |||
'nohup' allows you to logout from the terminal window without 'hangup' - otherwise, a forced closure of the window could kill the job. | |||
Output and errors normally sent to the terminal window are redirected to the OUT files. | |||
To check the job started as expected, try | |||
> head OUT | |||
precomputing function requisites... | |||
loading state... | |||
t : 199.999999999963 --> 0.000000000000000E+000 | |||
Re : 2400.00000000000 --> 2500.00000000000 | |||
N : 60 --> 64 | |||
initialising output files... | |||
timestepping..... | |||
Here we see that the state used as the initial condition was saved at simulation time 200, and that the time has been reset to 0. We also see the increase in Reynolds number Re and a slight increase in the radial resolution, N. | |||
If a message like the following appears almost immediately in the terminal window | |||
'[1]+ Done nohup ./main.out ...' | |||
then it is likely that there was an error. In that case, try | |||
> less OUT | |||
> less OUT.err | |||
If there is a message about MPI libraries, and earlier you changed <tt>_Np</tt> from another value to 1, then you could try | |||
running instead with | |||
> mpirun -np 1 ./main.out > OUT 2> OUT.err & | |||
You might need to include a path to mpirun; copy the path from that to the mpif90 command in your <tt>Makefile</tt>. | |||
=== Monitor the run === | |||
Let the code run until new files are produced (~10mins): | |||
> ls -l | |||
-rw-rw-r-- 1 20 Dec 20 11:11 HOST | |||
-rw-rw-r-- 1 1083 Dec 20 11:11 main.info | |||
-rwxrwxr-x 1 4295089 Dec 20 11:11 main.out | |||
-rw-rw-r-- 1 16180 Dec 20 11:22 OUT | |||
-rw-rw-r-- 1 0 Dec 20 11:11 OUT.err | |||
-rw-rw-r-- 1 106 Dec 20 11:11 RUNNING | |||
-rw-rw-r-- 1 3389548 Dec 20 11:11 state0000.cdf.dat | |||
-rw-rw-r-- 1 3389548 Dec 20 11:16 state0001.cdf.dat | |||
-rw-rw-r-- 1 3389548 Dec 20 11:21 state0002.cdf.dat | |||
-rw-rw-r-- 1 3177740 Jun 10 2013 state.cdf.in | |||
-rw-rw-r-- 1 23655 Dec 20 11:22 tim_step.dat | |||
-rw-rw-r-- 1 33615 Dec 20 11:22 vel_energy.dat | |||
-rw-rw-r-- 1 33615 Dec 20 11:22 vel_friction.dat | |||
-rw-rw-r-- 1 2668 Dec 20 11:11 vel_prof0000.dat | |||
-rw-rw-r-- 1 2668 Dec 20 11:16 vel_prof0001.dat | |||
-rw-rw-r-- 1 2668 Dec 20 11:21 vel_prof0002.dat | |||
-rw-rw-r-- 1 2901 Dec 20 11:11 vel_spec0000.dat | |||
-rw-rw-r-- 1 2901 Dec 20 11:16 vel_spec0001.dat | |||
-rw-rw-r-- 1 2901 Dec 20 11:21 vel_spec0002.dat | |||
-rw-rw-r-- 1 33615 Dec 20 11:22 vel_totEID.dat | |||
The code outputs | |||
* '''snapshot data''', which includes a 4-digit number e.g. <tt>vel_spec0003.dat</tt> and <tt>state0012.cdf.dat</tt>, saved every <tt>i_save_rate1</tt> (typically 2000) timesteps, and | |||
* '''time-series data''', e.g. the energy as a function of time <tt>vel_energy.dat</tt>, saved every <tt>i_save_rate2</tt> (typically 10) timesteps . | |||
To see how far it has run, try | |||
> tail OUT | |||
step= 4050 its= 1 | |||
step= 4060 its= 1 | |||
step= 4070 its= 1 | |||
step= 4080 its= 1 | |||
step= 4090 its= 1 | |||
step= 4100 its= 1 | |||
Or | |||
> tail vel_energy.dat | |||
0.405000000000E+02 0.245265514603E+00 0.220063087694E+00 0.183078661054E+00 | |||
0.406000000000E+02 0.245218154814E+00 0.220035747234E+00 0.183089213146E+00 | |||
0.407000000000E+02 0.245172273310E+00 0.220011449291E+00 0.183099918474E+00 | |||
0.408000000000E+02 0.245127855329E+00 0.219990209680E+00 0.183110740101E+00 | |||
0.409000000000E+02 0.245084886492E+00 0.219972040124E+00 0.183121641848E+00 | |||
0.410000000000E+02 0.245043352978E+00 0.219956948128E+00 0.183132588846E+00 | |||
Sometimes the time-series files are buffered, i.e. no output is written until the buffer is filled, so don't panic if it doesn't seem to be updating. The OUT file is less likely to be buffered. | |||
Column 1 of <tt>vel_energy.dat</tt> is the time, column 2 is the energy in the perturbation to the mean flow. Columns 3 and 4 are the energies in the axial mean (k=0) and azimuthal mean (m=0) components respectively. | |||
Let the code run for a few minutes, give it a chance to save a few state files. | |||
To see the simulation times '''when state were saved''', | |||
> grep state OUT | less | |||
loading state... | |||
saving state0000 t= 0.000000000000000E+000 | |||
saving state0001 t= 20.0000000000003 | |||
saving state0002 t= 40.0000000000006 | |||
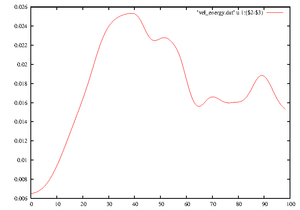
[[File:Tut_E3d.png|thumb|right|Energy vs time]] | |||
Let's plot the '''energy as a function of time''': | |||
<!-- need pre to preserve dollar symbol --> | |||
<pre> | |||
> gnuplot | |||
> plot 'vel_energy.dat' w l [with lines] | |||
> plot 'vel_energy.dat' u 1:($2-$3) w l [using column1 and column2-column3] | |||
</pre> | |||
The last line here plots the energy in the axially dependent modes only (k non-zero). This quantity decays rapidly after relaminarisations, and the simulation will stop if it drops below the parameter value <tt>d_minE3d</tt>, e.g. <tt>1d-5</tt>. Note that this parameter has no effect if it is set to e.g. <tt>-1d0</tt>. | |||
[[File:Tut_spec0001.png|thumb|right|Spectrum]] | |||
It is a good idea to keep track of the '''resolution'''. Still in <tt>gnuplot</tt> | |||
<pre> | |||
> set log | |||
> plot [][1e-6:] 'vel_spec0001.dat' u ($1+1):2 w lp [with lines and points] | |||
... | |||
> quit | |||
</pre> | |||
This is a rough plot indicating the drop-off in amplitude of coefficients, defined by E_k = max_{nm} a_{nkm}, E_m = max_{nk} a_{nkm}, E_n = max_{km} a_{nkm}, where n is the index of axial resolution, k for axial and m for azimuthal. | |||
Ideally, the lines should all drop by around 3-4 orders of magnitude or more. If there is an upward spike at the end of one of the lines, then this is usually the signature of a timestep that is too large. | |||
Ignore the zig-zag for the tail of the line for radial resolution, focus on the upper values in the zig-zag only. It arises because a finite difference scheme is used, but data has been transformed onto a spectral basis, purely for the purpose of gauging the quality of resolution. | |||
=== End the run === | |||
Type | |||
> rm RUNNING | |||
and press enter. This signals to the job to terminate (cleanly). | |||
Wait a few seconds then press enter again. There should be a message like, | |||
'<tt>[1]+ Done nohup ./main.out ...</tt>', to say that the job has has ended. | |||
To see how long and how fast it was running, do | |||
> tail OUT | |||
step= 9770 its= 1 | |||
step= 9780 its= 1 | |||
step= 9790 its= 1 | |||
step= 9800 its= 1 | |||
RUNNING deleted ! | |||
cleanup... | |||
sec/step = 0.1538023 | |||
CPU time = 25 mins. | |||
saving state0005 t= 98.1000000000133 | |||
...done! | |||
=== Make a util === | |||
Here we'll build a utility code to process data for visualisation. | |||
Changing the core code in <tt>program/</tt> should be avoided; almost any post-processing and runtime processing can be done by creating a utility instead. There are many code examples in the <tt>utils/</tt> directory. Further information can be found on the [[Utilities]] page. | |||
Let's return to the code, | |||
cd <path>/openpipeflow-x.xx/ | |||
Let's compile the utility | |||
utils/prim2matlab.f90 | |||
In <tt>Makefile</tt>, ensure that it is set to make the <tt>prim2matlab</tt> utility, | |||
UTIL = prim2matlab [ommit the .f90 extension] | |||
Now, | |||
> make | |||
> make install | |||
> make util | |||
Really, only the last command is necessary, which creates the executable <tt>prim2matlab.out</tt>. | |||
It is good practice, however, to do the previous commands to generate a [[main.info]] file to keep alongside the executable. | |||
=== Visualise === | |||
Fourier coefficients are stored in the state files. (It takes much less space and is more convenient should resolution be changed.) | |||
The executable <tt>prim2matlab.out</tt> converts data and outputs it to a NetCDF file that is easily read by Matlab and Visit software packages. It can be used to create cross sections, calculate functions on a volume to create isosurfaces, etc. | |||
Output is written to the file <tt>mat_vec.cdf</tt>: | |||
* contains variables A (the data) and x,y,z (grid points). | |||
* x has dimension x(nx). | |||
* A has dimension A(nx,ny,nz,Ads). | |||
* Ads=1 for scalar data, Ads=3 for a vector. | |||
* nz=1 if data is confined to a cross section. | |||
This section is to be extended. | |||
For the moment, please see scripts that accompany the code in the directory <tt>matlab/</tt>, in particular the file '''<tt>matlab/Readme.txt</tt>''' . | |||
Latest revision as of 01:52, 15 July 2020
This tutorial aims to guide a new user through
- setting up parameters for a job,
- monitoring a job's progress,
- simple plotting of time series from a run,
- visualisation of structures in a snapshot,
- manipulating data.
It is advised that you read through the Getting_started before trying the tutorial, but you may skip the Libraries section if someone has set up the libraries and Makefile for you.
The following assumes that the command 'make' does not exit with an error, i.e. that the code has been downloaded (download), and that libraries have been correctly installed (Getting_started#Compiling_libraries).
Overview
Where to start - initial conditions and the main.info file
The best input initial condition, state.cdf.in is usually the output state from another run, preferably from a run with similar parameter settings. Output state files are named state0000.cdf.dat, state0001.cdf.dat, state0002.cdf.dat, and so on. Any of these could be used as a potential initial condition. If resolution parameters do not match, then they automatically interpolated or truncated to the new resolution (the resolution selected at compile time).
In this example we'll use a travelling wave solution at Re=2400 as an initial condition for a run at Re=2500. Download the following file from the Database: File:Re2400a1.25.tgz and extract the contents:
> tar -xvvzf Re2400a1.25.tgz > cd Re2400a1.25/ > ls -l -rw-rw-r-- 1 758 Jul 16 2014 main.info -rw-rw-r-- 1 3177740 Jun 10 2013 state0010.cdf.dat
The directory Re2400a1.25/ contains an output state file state0010.cdf.dat and a main.info file. The main.info file is a plain text record of parameter settings that were used when compiling the executable that produced the state file.
A more computationally demanding example: You may wish to start from File:Re5300.Retau180.5D.tgz and compare data with Eggels et al. (1994) JFM. To run at a reasonable pace, try running on 5-10 cores.
Set your parameters
In a terminal window, take a look at the main.info file downloaded a moment ago
> less Re2400a1.25/main.info
('less' is like 'more', but you can scroll and search (with '/'). Press 'q' to exit.) The main.info file records the parameters associated with the state file, which will be used as the initial condition.
In another terminal window, edit the parameters for the run, so that they are the same as in the given main.info file, except with Re=2500. (If you have not yet changed the parameters.f90 file, it'll not need editing.)
> nano program/parameters.f90 [OR] > pico program/parameters.f90 [OR] > gedit program/parameters.f90
In the parameters.f90 file, you should ignore from where i_KL is declared onwards.
We will assume serial use (for parallel use see Getting_started#Typical_usage). The number of cores is set in parallel.h. Ensure that _Nr and _Ns are both defined to be 1 (number of cores _Np=_Nr*_Ns):
> head parallel.h ... #define _Nr 1 #define _Ns 1 ...
If not, edit with your favourite text editor, e.g.
> nano parallel.h
and set _Nr and _Ns as above.
Compile and setup a job directory
After setting the parameters, we need to create an executable that will run with the settings we've chosen. To compile the code with the current parameter settings
> make > make install
If an error is produced, see the top of this page - check that libraries and Makefile are set up correctly. The second command creates the directory install/ and a new main.info file. Often its a good idea to check for any differences in parameters between jobs:
> diff install/main.info Re2400a1.25/main.info
In this case the important difference is Re=2400 increased to Re=2500.
We'll create a new job directory with an initial condition in there ready for the new run
> cp Re2400a1.25/state0010.cdf.dat install/state.cdf.in > mkdir ~/runs/ > mv install ~/runs/job0001 > cd ~/runs/job0001 > ls -l -rw-rw-r-- 1 949 Sep 9 08:50 main.info -rw-rw-r-- 1 3679640 Sep 9 08:50 main.out -rw-rw-r-- 1 3177740 Jun 10 2013 state.cdf.in
Start the run
To start the run
> nohup ./main.out > OUT 2> OUT.err &
'&' puts the job in the background. 'nohup' allows you to logout from the terminal window without 'hangup' - otherwise, a forced closure of the window could kill the job. Output and errors normally sent to the terminal window are redirected to the OUT files.
To check the job started as expected, try
> head OUT precomputing function requisites... loading state... t : 199.999999999963 --> 0.000000000000000E+000 Re : 2400.00000000000 --> 2500.00000000000 N : 60 --> 64 initialising output files... timestepping.....
Here we see that the state used as the initial condition was saved at simulation time 200, and that the time has been reset to 0. We also see the increase in Reynolds number Re and a slight increase in the radial resolution, N.
If a message like the following appears almost immediately in the terminal window
'[1]+ Done nohup ./main.out ...'
then it is likely that there was an error. In that case, try
> less OUT > less OUT.err
If there is a message about MPI libraries, and earlier you changed _Np from another value to 1, then you could try running instead with
> mpirun -np 1 ./main.out > OUT 2> OUT.err &
You might need to include a path to mpirun; copy the path from that to the mpif90 command in your Makefile.
Monitor the run
Let the code run until new files are produced (~10mins):
> ls -l -rw-rw-r-- 1 20 Dec 20 11:11 HOST -rw-rw-r-- 1 1083 Dec 20 11:11 main.info -rwxrwxr-x 1 4295089 Dec 20 11:11 main.out -rw-rw-r-- 1 16180 Dec 20 11:22 OUT -rw-rw-r-- 1 0 Dec 20 11:11 OUT.err -rw-rw-r-- 1 106 Dec 20 11:11 RUNNING -rw-rw-r-- 1 3389548 Dec 20 11:11 state0000.cdf.dat -rw-rw-r-- 1 3389548 Dec 20 11:16 state0001.cdf.dat -rw-rw-r-- 1 3389548 Dec 20 11:21 state0002.cdf.dat -rw-rw-r-- 1 3177740 Jun 10 2013 state.cdf.in -rw-rw-r-- 1 23655 Dec 20 11:22 tim_step.dat -rw-rw-r-- 1 33615 Dec 20 11:22 vel_energy.dat -rw-rw-r-- 1 33615 Dec 20 11:22 vel_friction.dat -rw-rw-r-- 1 2668 Dec 20 11:11 vel_prof0000.dat -rw-rw-r-- 1 2668 Dec 20 11:16 vel_prof0001.dat -rw-rw-r-- 1 2668 Dec 20 11:21 vel_prof0002.dat -rw-rw-r-- 1 2901 Dec 20 11:11 vel_spec0000.dat -rw-rw-r-- 1 2901 Dec 20 11:16 vel_spec0001.dat -rw-rw-r-- 1 2901 Dec 20 11:21 vel_spec0002.dat -rw-rw-r-- 1 33615 Dec 20 11:22 vel_totEID.dat
The code outputs
- snapshot data, which includes a 4-digit number e.g. vel_spec0003.dat and state0012.cdf.dat, saved every i_save_rate1 (typically 2000) timesteps, and
- time-series data, e.g. the energy as a function of time vel_energy.dat, saved every i_save_rate2 (typically 10) timesteps .
To see how far it has run, try
> tail OUT step= 4050 its= 1 step= 4060 its= 1 step= 4070 its= 1 step= 4080 its= 1 step= 4090 its= 1 step= 4100 its= 1
Or
> tail vel_energy.dat 0.405000000000E+02 0.245265514603E+00 0.220063087694E+00 0.183078661054E+00 0.406000000000E+02 0.245218154814E+00 0.220035747234E+00 0.183089213146E+00 0.407000000000E+02 0.245172273310E+00 0.220011449291E+00 0.183099918474E+00 0.408000000000E+02 0.245127855329E+00 0.219990209680E+00 0.183110740101E+00 0.409000000000E+02 0.245084886492E+00 0.219972040124E+00 0.183121641848E+00 0.410000000000E+02 0.245043352978E+00 0.219956948128E+00 0.183132588846E+00
Sometimes the time-series files are buffered, i.e. no output is written until the buffer is filled, so don't panic if it doesn't seem to be updating. The OUT file is less likely to be buffered.
Column 1 of vel_energy.dat is the time, column 2 is the energy in the perturbation to the mean flow. Columns 3 and 4 are the energies in the axial mean (k=0) and azimuthal mean (m=0) components respectively.
Let the code run for a few minutes, give it a chance to save a few state files. To see the simulation times when state were saved,
> grep state OUT | less loading state... saving state0000 t= 0.000000000000000E+000 saving state0001 t= 20.0000000000003 saving state0002 t= 40.0000000000006
Let's plot the energy as a function of time:
> gnuplot > plot 'vel_energy.dat' w l [with lines] > plot 'vel_energy.dat' u 1:($2-$3) w l [using column1 and column2-column3]
The last line here plots the energy in the axially dependent modes only (k non-zero). This quantity decays rapidly after relaminarisations, and the simulation will stop if it drops below the parameter value d_minE3d, e.g. 1d-5. Note that this parameter has no effect if it is set to e.g. -1d0.
It is a good idea to keep track of the resolution. Still in gnuplot
> set log > plot [][1e-6:] 'vel_spec0001.dat' u ($1+1):2 w lp [with lines and points] ... > quit
This is a rough plot indicating the drop-off in amplitude of coefficients, defined by E_k = max_{nm} a_{nkm}, E_m = max_{nk} a_{nkm}, E_n = max_{km} a_{nkm}, where n is the index of axial resolution, k for axial and m for azimuthal.
Ideally, the lines should all drop by around 3-4 orders of magnitude or more. If there is an upward spike at the end of one of the lines, then this is usually the signature of a timestep that is too large.
Ignore the zig-zag for the tail of the line for radial resolution, focus on the upper values in the zig-zag only. It arises because a finite difference scheme is used, but data has been transformed onto a spectral basis, purely for the purpose of gauging the quality of resolution.
End the run
Type
> rm RUNNING
and press enter. This signals to the job to terminate (cleanly). Wait a few seconds then press enter again. There should be a message like, '[1]+ Done nohup ./main.out ...', to say that the job has has ended.
To see how long and how fast it was running, do
> tail OUT step= 9770 its= 1 step= 9780 its= 1 step= 9790 its= 1 step= 9800 its= 1 RUNNING deleted ! cleanup... sec/step = 0.1538023 CPU time = 25 mins. saving state0005 t= 98.1000000000133 ...done!
Make a util
Here we'll build a utility code to process data for visualisation.
Changing the core code in program/ should be avoided; almost any post-processing and runtime processing can be done by creating a utility instead. There are many code examples in the utils/ directory. Further information can be found on the Utilities page.
Let's return to the code,
cd <path>/openpipeflow-x.xx/
Let's compile the utility
utils/prim2matlab.f90
In Makefile, ensure that it is set to make the prim2matlab utility,
UTIL = prim2matlab [ommit the .f90 extension]
Now,
> make > make install > make util
Really, only the last command is necessary, which creates the executable prim2matlab.out. It is good practice, however, to do the previous commands to generate a main.info file to keep alongside the executable.
Visualise
Fourier coefficients are stored in the state files. (It takes much less space and is more convenient should resolution be changed.) The executable prim2matlab.out converts data and outputs it to a NetCDF file that is easily read by Matlab and Visit software packages. It can be used to create cross sections, calculate functions on a volume to create isosurfaces, etc.
Output is written to the file mat_vec.cdf:
- contains variables A (the data) and x,y,z (grid points).
- x has dimension x(nx).
- A has dimension A(nx,ny,nz,Ads).
- Ads=1 for scalar data, Ads=3 for a vector.
- nz=1 if data is confined to a cross section.
This section is to be extended. For the moment, please see scripts that accompany the code in the directory matlab/, in particular the file matlab/Readme.txt .